Specifying the grid designation process
The workflow for simulating a biodiversity data cube used in gcube can be divided in three steps or processes:
- Occurrence process
- Detection process
- Grid designation process
This tutorial documents the third part of the gcube simulation workflow, viz. the grid designation process.
# Load packageslibrary(gcube)
library(sf) # work with spatial objectslibrary(dplyr) # data wranglinglibrary(ggplot2) # data visualisationlibrary(ggExtra) # enhance data visualisationInput
The functions are set up such that a single polygon as input is enough to go through this workflow using default arguments.
The user can change these arguments to allow for more flexibility. As input, we create a polygon in which we simulate occurrences. It represents the spatial extent of the species.
polygon <- st_polygon(list(cbind(c(500, 1000, 1000, 600, 200, 100, 500), c(200, 100, 700, 1000, 900, 500, 200))))The polygon looks like this.
ggplot() + geom_sf(data = polygon) + theme_minimal()
Also consider a road across our polygon.
# Define the road widthroad_width <- 50
# Create road pointsroad_points <- rbind(c(100, 500), c(1000, 500))
# Create road-like polygon within the given polygonroad_polygon <- st_linestring(road_points) %>% st_buffer(road_width) %>% st_intersection(polygon) %>% st_polygon() %>% st_sfc() %>% st_as_sf() %>% rename(geometry = x)The result looks like this.
ggplot() + geom_sf(data = polygon, fill = "lightgreen") + geom_sf(data = road_polygon) + theme_minimal()
We can for example sample randomly within the polygon over 6 time points were we use a random walk over time with an initial average number of occurrences equal to 100 (see tutorial 1 about simulating the occurrence process).
occurrences_df <- simulate_occurrences( species_range = polygon, initial_average_occurrences = 100, n_time_points = 6, temporal_function = simulate_random_walk, sd_step = 1, spatial_pattern = "random", seed = 123)#> [using unconditional Gaussian simulation]This is the spatial distribution of the occurrences for each time point
ggplot() + geom_sf(data = polygon) + geom_sf(data = occurrences_df) + facet_wrap(~time_point, nrow = 2) + ggtitle("Distribution of occurrences for each time point") + theme_minimal()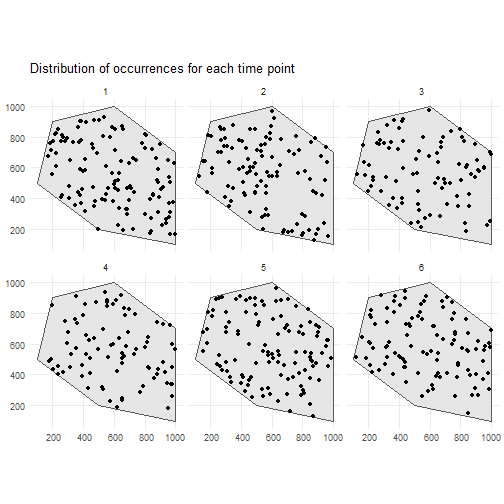
We detect occurrences using a 0.9 detection probability and a bias of 0.1 to detect occurrences on the road (see tutorial 2 about simulating the detection process).
detections_df_raw <- sample_observations( occurrences_df, detection_probability = 0.9, sampling_bias = "polygon", bias_area = road_polygon, bias_strength = 0.1, seed = 123)This is the spatial distribution of the occurrences for each time point
ggplot() + geom_sf(data = polygon, fill = "lightgreen") + geom_sf(data = road_polygon) + geom_sf(data = detections_df_raw, aes(colour = observed)) + scale_colour_manual(values = c("blue", "red")) + facet_wrap(~time_point, nrow = 2) + labs(title = "Distribution of occurrences for each time point") + theme_minimal()
We only keep the detected occurrences and add 25 meters of uncertainty to each observation (see tutorial 2 about simulating the detection process).
# Keep detected occurrencesdetections_df <- filter_observations( observations_total = detections_df_raw)
# Add 25 m coordinate uncertaintyobservations_df <- add_coordinate_uncertainty( observations = detections_df, coords_uncertainty_meters = 25)The final observations with uncertainty circles look like this.
# Create sf object with uncertainty circlesbuffered_observations <- st_buffer( observations_df, observations_df$coordinateUncertaintyInMeters)
# Visualiseggplot() + geom_sf(data = polygon, fill = "lightgreen") + geom_sf(data = road_polygon) + geom_sf(data = buffered_observations, fill = alpha("firebrick", 0.3)) + geom_sf(data = observations_df, colour = "firebrick", size = 0.8) + facet_wrap(~time_point, nrow = 2) + labs(title = "Distribution of observations for each time point") + theme_minimal()
Grid designation
Now we can make a data cube from our observations while taking into account the uncertainty.
We can create the grid using the grid_designation() function.
?grid_designationWe also need a grid. Each observation will be designated to a grid cell.
cube_grid <- st_make_grid( st_buffer(polygon, 25), n = c(20, 20), square = TRUE) %>% st_sf()The grid looks like this.
ggplot() + geom_sf(data = polygon) + geom_sf(data = cube_grid, alpha = 0) + theme_minimal()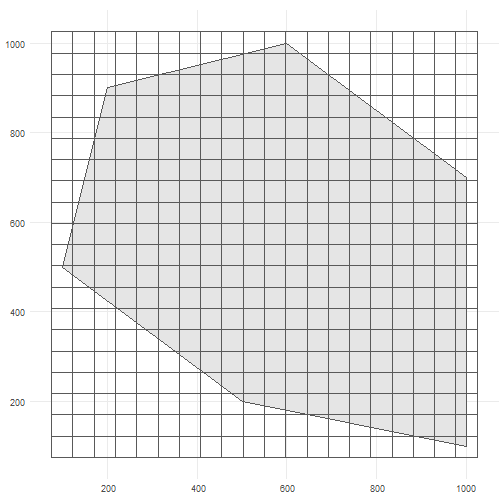
How does grid designation take coordinate uncertainty into account?
The default is "uniform" randomisation where a random point within the uncertainty circle is taken as the location of the observation.
This point is then designated to the overlapping grid cell.
Another option is "normal" where a point is sampled from a bivariate Normal distribution with means equal to the observation point and the variance equal to (-coordinateUncertaintyInMeters^2) / (2 * log(1 - p_norm)) such that p_norm % of all possible samples from this Normal distribution fall within the uncertainty circle.
This can be visualised by using these supporting functions.
?sample_from_uniform_circle?sample_from_binormal_circleLets create a single random point with 25 meter coordinate uncertainty. We sample 1000 times using uniform and normal randomisation to look at the difference between the methods.
# Create point and add coordinate uncertaintypoint_df <- tibble( x = 200, y = 500, time_point = 1, coordinateUncertaintyInMeters = 25) %>% st_as_sf(coords = c("x", "y"))
# Number of simulationsn_sim <- 1000We take 1000 samples with uniform randomisation.
list_samples_uniform <- vector("list", length = n_sim)for (i in seq_len(n_sim)) { sampled_point_uniform <- sample_from_uniform_circle(point_df) sampled_point_uniform$sim <- i list_samples_uniform[[i]] <- sampled_point_uniform}samples_uniform_df <- do.call(rbind.data.frame, list_samples_uniform)We take 1000 samples with normal randomisation
list_samples_normal <- vector("list", length = n_sim)for (i in seq_len(n_sim)) { sampled_point_normal <- sample_from_binormal_circle(point_df, p_norm = 0.95) sampled_point_normal$sim <- i list_samples_normal[[i]] <- sampled_point_normal}samples_normal_df <- do.call(rbind.data.frame, list_samples_normal)# Get coordinatescoordinates_uniform_df <- data.frame(st_coordinates(samples_uniform_df))coordinates_normal_df <- data.frame(st_coordinates(samples_normal_df))coordinates_point_df <- data.frame(st_coordinates(point_df))
# Create figures for both randomisationsscatter_uniform <- ggplot() + geom_point(data = coordinates_uniform_df, aes(x = X, y = Y), colour = "cornflowerblue") + geom_segment(data = coordinates_point_df, aes(x = X, xend = X + 25, y = Y, yend = Y), linewidth = 1.5, colour = "darkgreen") + geom_label(aes(y = 503, x = 212.5, label = "25 m"), colour = "black", size = 5) + geom_point(data = coordinates_point_df, aes(x = X, y = Y), color = "firebrick", size = 2) + coord_fixed() + theme_minimal()
scatter_normal <- ggplot() + geom_point(data = coordinates_normal_df, aes(x = X, y = Y), colour = "cornflowerblue") + geom_segment(data = coordinates_point_df, aes(x = X, xend = X + 25, y = Y, yend = Y), linewidth = 1.5, colour = "darkgreen") + geom_label(aes(y = 503, x = 212.5, label = "25 m"), colour = "black", size = 5) + stat_ellipse(data = coordinates_normal_df, aes(x = X, y = Y), level = 0.975, linewidth = 1, color = "firebrick") + geom_point(data = coordinates_point_df, aes(x = X, y = Y), color = "firebrick", size = 2) + coord_fixed() + theme_minimal()In the case of uniform randomisation, we see samples everywhere and evenly spread within the uncertainty circle.
ggExtra::ggMarginal(scatter_uniform, type = "histogram")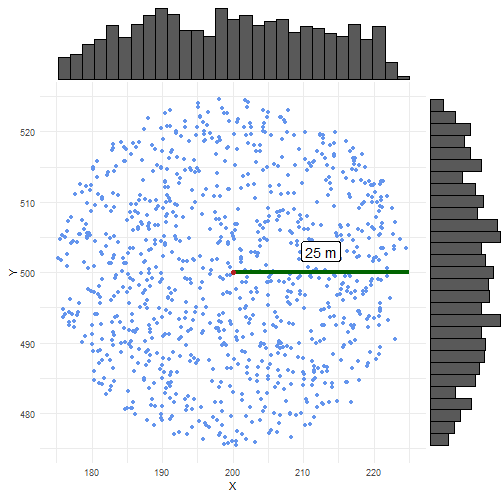
In the case of normal randomisation, we see some samples outside the uncertainty circle.
This should be 0.05 (=1 - p_norm) %.
We also see more samples closer to the central point.
ggExtra::ggMarginal(scatter_normal, type = "histogram")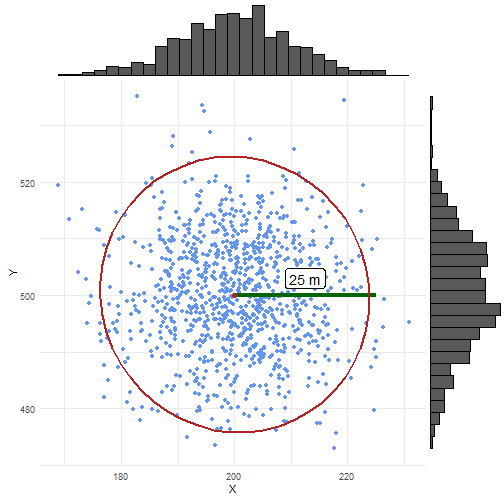
If no coordinate uncertainty is provided, the original observation point is used for grid designation.
Example
Now we know how to use the randomisation in grid_designation().
By default we use uniform randomisation.
We create an occurrence cube for time point 1.
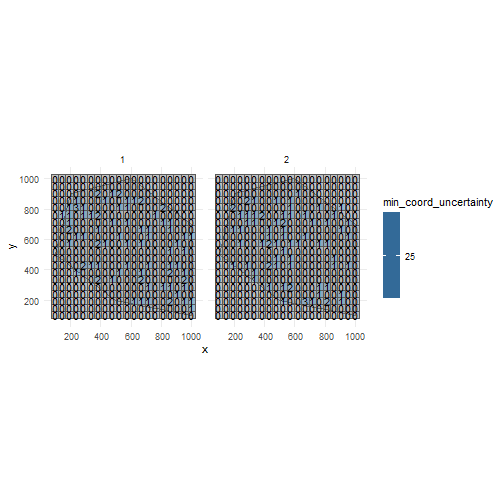
occurrence_cube_df <- grid_designation( observations_df, cube_grid, seed = 123)For each grid cell (column cell_code) at each time point (column time_point), we get the number of observations (column n, sampled within uncertainty circle) and the minimal coordinate uncertainty (column min_coord_uncertainty).
The latter is 25 for each grid cell since each observation had the same coordinate uncertainty.
head(occurrence_cube_df %>% st_drop_geometry())#> # A tibble: 6 × 4#> time_point cell_code n min_coord_uncertainty#> <int> <chr> <int> <dbl>#> 1 1 107 2 25#> 2 1 109 1 25#> 3 1 113 2 25#> 4 1 119 2 25#> 5 1 124 1 25#> 6 1 130 1 25Get sampled points within uncertainty circle by setting aggregate = FALSE.
sampled_points <- grid_designation( observations_df, cube_grid, seed = 123, aggregate = FALSE)Lets visualise were the samples were taken for time point 1.
Note that no distinction is made between zeroes and NA values!
Every absence gets a zero value.
ggplot() + geom_sf(data = polygon) + geom_sf(data = occurrence_cube_df %>% dplyr::filter(time_point == 1), alpha = 0) + geom_sf_text(data = occurrence_cube_df %>% dplyr::filter(time_point == 1), aes(label = n)) + geom_sf(data = buffered_observations %>% dplyr::filter(time_point == 1), fill = alpha("firebrick", 0.3)) + geom_sf(data = sampled_points %>% dplyr::filter(time_point == 1), colour = "blue") + geom_sf(data = observations_df %>% dplyr::filter(time_point == 1), colour = "firebrick") + theme_minimal()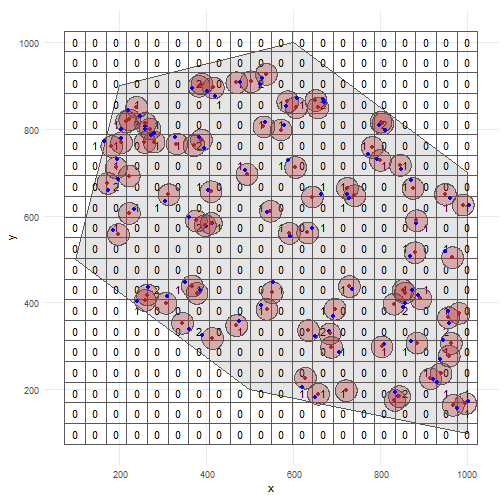
Visualise minimal coordinate uncertainty for time points 1 and 2.
ggplot() + geom_sf(data = polygon) + geom_sf(data = occurrence_cube_df %>% dplyr::filter(time_point %in% 1:2), aes(fill = min_coord_uncertainty), alpha = 0.5) + geom_sf_text(data = occurrence_cube_df %>% dplyr::filter(time_point %in% 1:2), aes(label = n)) + facet_wrap(~time_point) + theme_minimal()